Microarrays
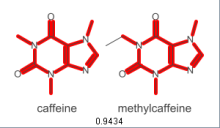
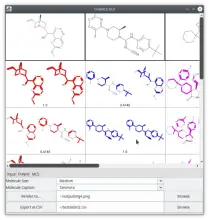
One tool I created recently, enables one to look at microarray-like data and highlight genes which are hypothesised to have a significant difference from the norm. You can use this on CRISPR data for example.
The main highlight of this its ability to be containerised as a docker image, and thus being easy to install and otherwise deployable on any machine as a web interface. Thus, it should simply be a case of giving it the data you want (format permitting), running the app and seeing the output in a web browser.
See the GIT link below for more information.